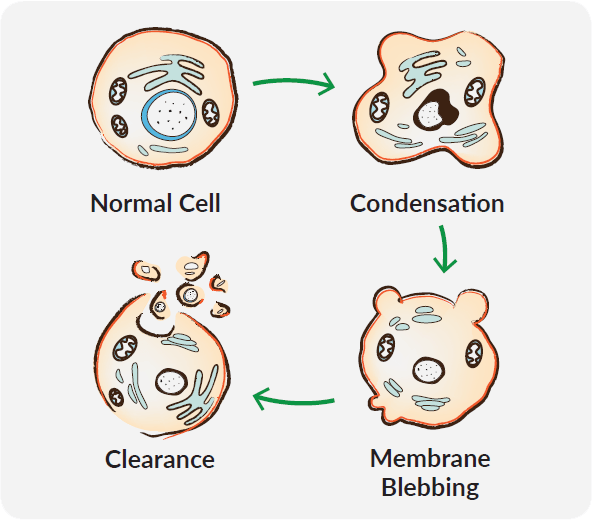
Apoptosis encompasses a non-inflammatory cascade of molecular events and results in distinct morphological changes including reduction in cellular volume, nuclear fragmentation, condensation of chromatin, DNA degradation, cell membrane blebbing, and subsequent dispersal into apoptotic bodies. This sequence not only contributes to the destruction of unwanted cells but prepares their debris for contained removal by phagocytes. Due to the variety of apoptotic stimuli and subtleties in resulting pathways, it can be critical to determine which correlates of cell death are affected by your experimental conditions.
Mitochondrial Membrane Potential
Mitochondria are intricately involved in the apoptotic process at several levels. Throughout the life of a cell, mitochondria use oxidizable substrates to produce a proton gradient across the mitochondrial inner membrane. During apoptosis, this transmembrane potential decreases in association with the opening of the mitochondrial permeability pore and the release of apoptogenic factors, such as cytochrome c. In some apoptotic models, loss of membrane potential is thought to be an early event in the apoptotic process. However, others suggest that loss of membrane potential is a consequence of the apoptotic signaling pathway.2-4 A simple way to assess membrane potential of mitochondria in a cell population is the use of positively charged dyes, such as JC-1 and TMRE, that accumulate in the electronegative interior of active mitochondria.
JC-1/TMRE
JC-1 changes from red fluorescence when aggregated in healthy mitochondria to green fluorescence when membrane potential is lost, as in apoptosis. TMRE accumulates in polarized mitochondria and is suitable for monitoring changes in mitochondrial membrane potential in live cells. Depolarized mitochondria with decreased membrane potential are unable to accumulate TMRE.
Caspase Activation
The main orchestrators of apoptotic signaling are a subset of Cysteine-dependent ASPartate proteASES (caspases), whose efficient activation defines the life or death decisions in the cell. Caspases are activated by several pathways and form feed-forward loops to cement apoptotic cell fate. The extrinsic pathway involves the binding of extracellular death ligands (such as FasL or TNF-α) to transmembrane death receptors, which recruit initiator caspase-8. In the intrinsic pathway, cytochrome c released from mitochondria can trigger the assembly of a caspase-activating complex (called the apoptosome) that recruits and activates initiator caspase-9. Initiator caspases cleave and activate executioner caspases-3 and -7, culminating in cleavage of specific substrates that directly result in the morphological changes classically defined as cellular apoptosis.5 Thus, measuring caspase activity serves as an important indicator of the ongoing apoptotic process.
Caspase-3/7
The protease activity of caspase-3 and -7 can be detected by using the sequence-specific fluorogenic substrate, N-Ac-DEVD-N'-MC-R110, which upon cleavage, generates a fluorescent product.
Nuclear Fragmentation, Chromatin Condensation, and DNA Degradation
Caspases coordinate the dismantling of internal cell structures, which ultimately aids in the efficiency of their disposal. One of the most noticeable and distinctive events of this process is the condensation of the nucleus and its fragmentation into smaller pieces. Nuclear fragmentation results from the disintegration of the nuclear lamina via proteolysis by caspases and the collapse of the nuclear envelope. Another distinguishing characteristic of apoptosis is the condensation of chromatin accompanied by the hydrolysis of nuclear DNA into a ladder of fragments. Hoechst 33342, a cell-permeable, fluorescent nucleic acid stain, is often used to identify nuclear condensation by microscopic analysis. The Golgi, endoplasmic reticulum, and mitochondrial networks also undergo fragmentation during apoptosis. As they are dismantled, the pieces are dispersed into plasma membrane blebs.
DNA Stains
Hoechst dyes are cell-permeable and bind to nucleic acids in live or fixed cells. Their blue emission spectrum is convenient for researchers who plan to use multiple fluorescent stains in a single sample. An alternate cell-permeable DNA stain is DRAQ5™, whose far-red excitation can be multiplexed with fluorophores with blue to orange emission. Membrane impermeant dyes, such as propidium iodide, DAPI, DRAQ7™, or RedDot™2, are ideal for specifically staining the nuclei of dead cells, whose plasma membrane integrity is compromised.
Membrane Blebbing
Caspases also cleave many of the major constituents of the cell cytoskeleton, which contributes to the rounding and retraction of the cell that is seen in the early stages of apoptosis. An additional consequence of cytoskeleton weakening is dynamic membrane blebbing. As the cell cytoplasm pushes against unsupported areas of the plasma membrane, membrane protrusions can be visualized microscopically. This is thought to result from myosin-dependent contraction of cortical bundles of actin, pushing the cytosol against the cell cortex and causing blebs in areas where the cytoskeleton has deteriorated. Membrane blebbing is a key step toward apoptotic cells budding off into smaller apoptotic bodies.
Apoptotic Clearance Signals
A critical component of the apoptotic process is the distinct lack of an inflammatory response to dying cells. The collapse of apoptotic cells into apoptotic bodies both prevents release of damage-associated molecular patterns into the extracellular space and makes disposal easier for phagocytes. Early in apoptosis, apoptotic cells secrete "find-me" signals to recruit engulfing cells and display "eat-me" signals that engulfing cells can recognize. This select interaction between apoptotic cells and engulfing cells ensures specific targeting of dying cells for non-inflammatory clearance. "Find-me" signals that recruit phagocytes include lysophosphatidylcholine, sphingosine-1-phosphate, fractalkine, and nucleotides like ATP and UTP. Through caspase-mediated channel opening, plasma membrane pannexin 1 (PANX1) channels release nucleotide "find-me" signals to recruit phagocytes.6 Caspase-dependent PANX1 channel opening also allows the entry of a small group of fluorescent monomeric cyanine dyes including TO-PRO®-3. Thus, pannexin channel functioning can be taken advantage of to identify cells that are sending "find-me" signals.7
Once the phagocyte is attracted to the apoptotic cell, it recognizes phosphatidylserine and phosphatidylethanolamine residues redistributed from the inner to outer leaflet of the phospholipid bilayer that are now exposed on the cell surface. This membrane rearrangement—a well-recognized hallmark of apoptosis—designates the apoptotic cell as different from its viable counterparts and, thus, is a sign for phagocytes to eat the foreign cell. The phospholipid binding protein Annexin V adheres to phosphatidylserine residues exposed during apoptosis. It is used to probe for phosphatidylserine on the outer membrane of apoptotic cells in order to identify the "eat-me" phase of apoptosis involving the redistribution of membrane phospholipids.8,9
TO-PRO®-3
TO-PRO®-3 is a far-red fluorescent dye that enables detection of a very early event in the initiation of apoptosis. It enters cells through selective pannexin channels that are activated in a caspase-dependent manner early in the apoptotic process. Thus, cells that might be considered viable due to lack of detection of traditional apoptosis markers (i.e., Annexin V) can be identified as having just begun the apoptotic process.
Annexin V
Annexin V can be conjugated to various fluorochromes (e.g., FITC, PE, APC) to probe for phosphatidylserine on the outer membrane of apoptotic cells. Staining for Annexin V can be combined with markers of plasma membrane integrity (i.e., RedDot™2, propidium iodide, DAPI) to distinguish apoptotic cells from necrotic cells.
Multiplex is Best
These distinguishing morphological features and key parts of the apoptotic signaling pathway can be observed and measured to verify the mode of cell death in your model system. However, a single readout can be misleading under some experimental conditions (e.g., distinguishing apoptosis from necrosis). Multiplexed assays are available to provide a more complete understanding of the apoptotic process in a single sample. For example, measuring TO-PRO®-3, Annexin V, TMRE, and DAPI allows you to concomitantly evaluate "find-me" versus "eat-me" signals, mitochondrial membrane potential, and nuclear fragmentation for a more complex picture of how apoptosis is affecting your cells. Furthermore, with the discovery of early apoptosis markers (i.e., TO-PRO®-3), cells that might have been considered viable due to a lack of traditional apoptosis markers (i.e., Annexin V) can now be identified as having just begun the apoptotic process. Thus, a multiplexed method that includes multiple markers for various stages of apoptosis allows for the quantitative segmentation of the total sample population into distinct phases: viable, early apoptosis, late apoptosis, apoptotic bodies, and non-cellular debris.
The range of cell-based assays available from Cayman is designed to give you the flexibility and efficiency needed to determine the variable stages of cell death in your model system. Biomols technical support and Caymans product development scientists are here to help you choose the most appropriate cell-based assay for your experimental endpoint, so you can obtain the most accurate answers for your research questions.
References
- Hanahan, D. and Weinberg, R.A. Cell 144(5), 646-674 (2011).
- Gottlieb, E., Armour, S.M., Harris, M.H., et al. Cell Death Differ. 10(6), 709–717 (2003).
- Ly, J.D., Grubb, D.R., and Lawen, A. Apoptosis 8(2), 115-128 (2003).
- Vakifahmetoglu-Norberg, H., Ouchida, A.T., and Norberg, E. Biochem. Biophys. Res. Commun. 482(3), 426-431 (2017).
- Taylor, R.C., Cullen, S.P., and Martin, S.J. Nat. Rev. Mol. Cell Biol. 9(3), 231-241 (2008).
- Chekeni, F.B., Elliott, M.R., Sandilos, J.K., et al. Nature 467(7317), 863-867 (2010).
- Jiang, L., Tixeira, R., Caruso, S., et al. Nat. Protoc. 11(4), 655-663 (2016).
- van Engeland, M., Nieland, L.J., Ramaekers, F.C., et al. Cytometry 31(1), 1-9 (1998).
- Logue, S.E., Elgendy, M., and Martin, S.J. Nat. Protoc. 4(9), 1383-1395 (2009).