Cookie preferences
This website uses cookies, which are necessary for the technical operation of the website and are always set. Other cookies, which increase the comfort when using this website, are used for direct advertising or to facilitate interaction with other websites and social networks, are only set with your consent.
Configuration
Technically required
These cookies are necessary for the basic functions of the shop.
"Allow all cookies" cookie
"Decline all cookies" cookie
CSRF token
Cookie preferences
Currency change
Customer-specific caching
FACT-Finder tracking
Individual prices
Selected shop
Session
Comfort functions
These cookies are used to make the shopping experience even more appealing, for example for the recognition of the visitor.
Note
Show the facebook fanpage in the right blod sidebar
Statistics & Tracking
Affiliate program
Conversion and usertracking via Google Tag Manager
Track device being used
If you have any questions, please use our Contact Form.
You can also order by e-mail: info@biomol.com
Larger quantity required? Request bulk
You can also order by e-mail: info@biomol.com
Larger quantity required? Request bulk
Organism: Homo sapiens (Human). Source: E.coli. Expression Region: 1-639aa. Protein Length: Full... more
Product information "Poly (A)-specific ribonuclease PARN (PARN), human, recombinant"
Organism: Homo sapiens (Human). Source: E.coli. Expression Region: 1-639aa. Protein Length: Full Length. Tag Info: N-terminal 6xHis-tagged. Target Protein Sequence: MEIIRSNFKS NLHKVYQAIE EADFFAIDGE FSGISDGPSV SALTNGFDTP EERYQKLKKH SMDFLLFQFG LCTFKYDYTD SKYITKSFNF YVFPKPFNRS SPDVKFVCQS SSIDFLASQG FDFNKVFRNG IPYLNQEEER QLREQYDEKR SQANGAGALS YVSPNTSKCP VTIPEDQKKF IDQVVEKIED LLQSEENKNL DLEPCTGFQR KLIYQTLSWK YPKGIHVETL ETEKKERYIV ISKVDEEERK RREQQKHAKE QEELNDAVGF SRVIHAIANS GKLVIGHNML LDVMHTVHQF YCPLPADLSE FKEMTTCVFP RLLDTKLMAS TQPFKDIINN TSLAELEKRL KETPFNPPKV ESAEGFPSYD TASEQLHEAG YDAYITGLCF ISMANYLGSF LSPPKIHVSA RSKLIEPFFN KLFLMRVMDI PYLNLEGPDL QPKRDHVLHV TFPKEWKTSD LYQLFSAFGN IQISWIDDTS AFVSLSQPEQ VKIAVNTSKY AESYRIQTYA EYMGRKQEEK QIKRKWTEDS WKEADSKRLN PQCIPYTLQN HYYRNNSFTA PSTVGKRNLS PSQEEAGLED GVSGEISDTE LEQTDSCAEP LSEGRKKAKK LKRMKKELSP AGSISKNSPA TLFEVPDTW. Purity: Greater than 90% as determined by SDS-PAGE. Endotoxin: Not test. Biological Activity: n/a. Form: Liquid or Lyophilized powder. Buffer: If the delivery form is liquid, the default storage buffer is Tris/PBS-based buffer, 5%-50% glycerol. If the delivery form is lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer, 6% Trehalose, pH 8.0. Reconstitution: We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20 °C/-80 °C. Our default final concentration of glycerol is 50%. Customers could use it as reference. Storage: The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself. Generally, the shelf life of liquid form is 6 months at -20 °C/-80 °C. The shelf life of lyophilized form is 12 months at -20 °C/-80 °C. Notes: Repeated freezing and thawing is not recommended. Store working aliquots at 4 °C for up to one week. Relevance: 3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early bryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain prature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elents (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the roval of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization. Reference: The sequence and analysis of duplication-rich human chromosome 16.Martin J., Han C., Gordon L.A., Terry A., Prabhakar S., She X., Xie G., Hellsten U., Chan Y.M., Altherr M., Couronne O., Aerts A., Bajorek E., Black S., Blumer H., Branscomb E., Brown N.C., Bruno W.J. , Buckingham J.M., Callen D.F., Campbell C.S., Campbell M.L., Campbell E.W., Caoile C., Challacombe J.F., Chasteen L.A., Chertkov O., Chi H.C., Christensen M., Clark L.M., Cohn J.D., Denys M., Detter J.C., Dickson M., Dimitrijevic-Bussod M., Escobar J., Fawcett J.J., Flowers D., Fotopulos D., Glavina T., Gomez M., Gonzales E., Goodstein D., Goodwin L.A., Grady D.L., Grigoriev I., Groza M., Hammon N., Hawkins T., Haydu L., Hildebrand C.E., Huang W., Israni S., Jett J., Jewett P.B., Kadner K., Kimball H., Kobayashi A., Krawczyk M.-C., Leyba T., Longmire J.L., Lopez F., Lou Y., Lowry S., Ludeman T., Manohar C.F., Mark G.A., McMurray K.L., Meincke L.J., Morgan J., Moyzis R.K., Mundt M.O., Munk A.C., Nandkeshwar R.D., Pitluck S., Pollard M., Predki P., Parson-Quintana B., Ramirez L., Rash S., Retterer J., Ricke D.O., Robinson D.L., Rodriguez A., Salamov A., Saunders E.H., Scott D., Shough T., Stallings R.L., Stalvey M., Sutherland R.D., Tapia R., Tesmer J.G., Thayer N., Thompson L.S., Tice H., Torney D.C., Tran-Gyamfi M., Tsai M., Ulanovsky L.E., Ustaszewska A., Vo N., White P.S., Williams A.L., Wills P.L., Wu J.-R., Wu K., Yang J., DeJong P., Bruce D., Doggett N.A., Deaven L., Schmutz J., Grimwood J., Richardson P., Rokhsar D.S., Eichler E.E., Gilna P., Lucas S.M., Myers R.M., Rubin E.M., Pennacchio L.A.Nature 432:988-994(2004). Function: 3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization.
| Keywords: | DAN, PARN, EC=3.1.13.4, Deadenylation nuclease, Deadenylating nuclease, Poly(A)-specific ribonuclease PARN, Polyadenylate-specific ribonuclease, Recombinant Human Poly (A)-specific ribonuclease PARN (PARN) |
| Supplier: | Cusabio |
| Supplier-Nr: | EP017456HU |
Properties
| Application: | Activity not tested |
| Conjugate: | No |
| Host: | E.coli |
| Species reactivity: | human |
| MW: | 77.5 kD |
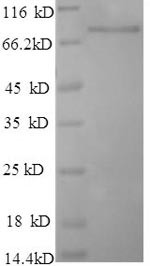
| Purity: | >90% (SDS-PAGE) |
Database Information
| KEGG ID : | K01148 | Matching products |
| UniProt ID : | O95453 | Matching products |
| Gene ID : | GeneID 5073 | Matching products |
Handling & Safety
| Storage: | -20°C |
| Shipping: | +4°C (International: +4°C) |
Caution
Our products are for laboratory research use only: Not for administration to humans!
Our products are for laboratory research use only: Not for administration to humans!
You will get a certificate here
Viewed

