Reviewed by Thomas G. Brock, Ph.D.
Scientists who are studying autoimmunity or rheumatoid arthritis know something that many others might not: citrullination of proteins is hugely important. This form of post-translational protein modification creates novel epitopes on common proteins, providing ‘neoantigens' that are now known to be important in autoimmune disease. Citrullination can occur outside of the cell, in the cytoplasm, and in the nucleus. Because of this, it can affect such diverse processes as blood clotting, skin differentiation, and chromatin remodeling. Although the roles for this process are only beginning to be understood, citrullination has established actions in inflammatory signaling. By the end of this article, readers should not only learn some hot science to impress friends but may even have new ideas for their research ventures.
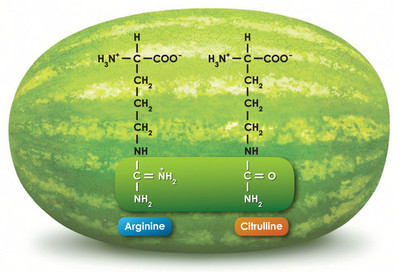
Citrulline and Watermelons
The biochemist should know about citrulline from the urea cycle, an important process for removing ammonia in animals. Citrulline is an α-amino acid that is structurally similar to arginine. In the urea cycle, carbamoyl phosphate is produced from ammonia, bicarbonate, and phosphate. Citrulline, synthesized from carbamoyl phosphate and ornithine by ornithine transcarbamylase, is simply an intermediate in the generation of arginine, which is then cleaved by arginase 1 to give urea and ornithine. In this cycle, citrulline is synthesized within the mitochondria and then exported to the cytoplasm for metabolism.
The serious gardener might recognize Citrullus lanatus (occasionally given, mistakenly, as C. vulgaris) as the formal name for watermelon. These cucurbits apparently originated in southern Africa and were cultivated in the Nile Valley some 4,000 years ago. Now grown worldwide in temperate climates, watermelon is remarkably rich in citrulline and arginine as free amino acids.1 In fact, the amino acid citrulline received its name when it was first isolated from the juice of watermelon over 80 years ago. As citrulline is a precursor of arginine and arginine can give rise to the vasoactive mediator nitric oxide, watermelon is not only tasty but also a potentially healthful treat.
Citrullination, PADs, and Rheumatoid Arthritis
Citrillunation is the enzymatic conversion of arginine to citrulline in situ on proteins (Figure 2). As the reaction involves the removal of an imine (double bonded nitrogen), it is catalyzed by deiminases. Known as peptidyl (protein) arginine deiminases (PAD, or PAD1), these enzymes are largely restricted to vertebrates, bacteria, and fungi. In human, rat, mouse, cow, horse, Rhesus monkey, and chimpanzee, there are 5 PAD genes, curiously numbered 1-4 and 6. In humans, each gene has a single gene product coding for a single, calcium-dependent enzyme, and each enzyme has unique substrates and functions. For example, PAD3 is found in the cytoplasm of hair follicles and keratinocytes, citrullinates filaggrin in follicles and trichohyalin in the inner root sheath, and regulates hair follicle and epidermal differentiation. Filaggrin and trichohyalin are intermediate filament-associated proteins that are involved in structurally aggregating or connecting keratin intermediate filaments. Similarly, some of the substrates of other PADs are structural proteins.2

Figure 2. PADs catalyze, Ca2+-dependent, imine to ketone reactions on specific peptidyl arginine side groups
Rheumatoid arthritis (RA) has long been considered to be a systemic autoimmune disease. A major breakthrough occurred in 1998 when Schellekens and colleagues reported that many RA autoantibodies detect citrulline on peptides, that many of these were specific for RA, and that the antibodies are present early in disease.3 Citrullination of specific proteins, including filaggrin, vimentin, fibrin, fibrinogen, α-enolase, and collagen II, produces novel epitopes that give rise to autoantibodies.4 While anti-citrullinated protein antibodies (ACPA) are now recognized as specific biomarkers for RA, assays that use cyclic citrullinated peptide (CCP) as an artificial antigen to probe for anti-CCP have high specificity and sensitivity.5,6 Importantly, the presence of anti-CCP antibodies can be predictive for erosive arthritis, suggesting that protein citrullination plays a role in the etiology of RA and that PAD inhibitors might be therapeutic.7
PAD4 is the principle citrullination enzyme in RA. It is expressed in neutrophils, eosinophils and tissue resident macrophages, but not lymphocytes or circulating monocytes. PAD4 normally has roles in cellular differentiation and the regulation of transcription of p53 and estrogen receptors. However, joint inflammation is characterized by infiltration of PAD4-bearing leukocytes. In one scenario, cellular activation, with a rise in intracellular calcium, leads to PAD4 citrullination of leukocyte vimentin, an intermediate filament involved in leukocyte attachment, migration, and cell signaling. Alternatively, widespread leukocyte necrosis resulting from release of cytotoxic compounds during degranulation results in the release of PAD4, as well as numerous cytoskeletal proteins, into the calcium-rich inflammatory milieu, resulting in indiscriminate citrullination of multiple targets. In either case, the stage is set for the development of autoantibodies. One question remains: why doesn't this happen in more inflammatory settings?
Citrullination Within the Nucleus
As mentioned above, each of the five PAD enzymes appears to have distinct substrates and roles (see Table 1). For example, while PAD1 and PAD3 are both associated with epithelium and can contribute to keratinocyte differentiation, PAD1 is primarily involved in maintaining cutaneous barrier function and PAD3 has a central role in follicle development. Of all the PADs, PAD4 appears to be unique in its ability to enter the nucleus. As a result, PAD4 can citrullinate nuclear substrates, including the histones H2A, H3, and H4, as well as the histone acetyltransferase p300. While the actions and implications of PAD4 in the nucleus are still poorly understood, interesting aspects have been revealed. First, deimination interplays with methylation of arginine residues: PAD4 can remove monomethyl groups from arginine residues through a demethyliminase activity, reversing the actions of protein arginine methyltransferases (PRMT) (Figure 3).8 PAD4 cannot, however, act on dimethylated residues. Similarly, deimination by PAD4 prevents arginine methylation.9 Thus, PAD4 can block or reverse some of the actions of PRMT.

Figure 3. PADs demethylate mono-methylated arginine residues (A) PADs produce citrullinated side chains on either normal arginine or arginine that has been mono-methylated by PRMTs. Aminotransferases (AT) may convert citrullinated chains to iminated chains. (B) PRMTs work with HATs to decondense chromatin. PADs and HDACs initiate the return of chromatin to a condensed form by citrullinating mono-methylated arginine and deacetylating lysine residues on histone tails.
PAD4 also interplays with protein acetylation/deacetylation. In order for some nuclear receptors to activate transcription in the presence of ligand, they must first recruit a p160 coactivator, which in turn binds the histone acetyltransferase p300. This allows acetylation that is necessary for chromatin remodeling, histone eviction, or transcriptional coactivation. The binding of p300 to the coactivator p160 is inhibited by methylation of Arg2142 on p300 by CARM1 (PRMT4), preventing transcription.10 This methylation mark is removed by PAD4, enhancing the p160-p300 interaction and transcription. In addition, PAD4 directly associates with histone deacetylase 1 (HDAC1), positioning the two proteins and their activities together in specific protein complexes.10 On the pS2 promoter, which is driven cyclically by the estrogen receptor alpha (ERα), PAD4 and HDAC1 bind and then release together in a periodic manner, with corresponding increases and decreases in citrullination and deacetylation of H3. The association of PAD4 and HDAC1 on pS2 produces a reversible, repressive chromatin environment.11,12
|
Table 1. PDA enzymes, their substrates and their cellular/tissue distributions |
||
| Enzyme | Substrates | Cellular/tissue distributions |
| PAD1 | keratin K1, filaggrin | epidermis, uterus, keratinocytes |
| PAD2 | vimentin, myelin basic protein, glial fibrillary acidic protein | skeletal muscle, brain, pancreas, glial cells, macrophages, bone marrow, breast, colon, embryo, eye, kidney, epidermis, uterus, thymus |
| PAD3 | Trichohyalin, filaggrin | hair follicles, keratinocytes |
| PAD4 | H2A, H3, H4, vimentin, nucleophosmin, p300, PAD4, 40S ribosomal protein S2, antithrombin | eosinophils, neutrophils |
| PAD5 | unknown | egg, ovary, early embryo, thymus, oocyte |
Cancer and Nets
The tumor suppressor protein p53 regulates the expression of various target genes, whose products in turn modulate cell growth and apoptosis. PAD4 and HDAC2 interact with p53 via distinct domains, resulting in their co-localization at p53-regulated promoters and suppression of transcription.13,14 The PAD4 inhibitor Cl-amidine and the HDAC inhibitor SAHA additively induce gene expression. PAD4's role as a transcription co-repressor for p53 may help explain the role of PAD4 in cancer: the deiminase is overexpressed in cancers of the breast, lung, liver, esophagus, colon, kidney, ovaries, endometrium, uterine, and bladder.15 The PAD4 inhibitors F- and Cl-amidine display cytotoxic effects towards cancerous cell lines but not against non-cancerous lines.16 This suggests that PAD4 inhibitors would be useful for treating certain cancers, as well as RA.
On a different front, citrullination by PAD4 plays a role in the immune response to bacteria. In addition to their vast granulocytic armamentarium, neutrophils unleash highly decondensed chromatin structures, called neutrophil extracellular traps (NETs) to trap and kill pathogenic bacteria.17 Impaired NET formation predisposes individuals to bacterial infection. Uncontrolled hypercitrullination of histones by PAD4 mediates chromatin decondensation.18 PAD4 deficient neutrophils cannot make NETs and PAD4 -/- mice are more susceptible to bacterial infection than PAD4 +/+ mice.19 Taken together, these results suggest that the systemic use of PAD4 inhibitors would compromise immune defense against infection.
References
- Tedesco, T.A., Benford, S.A., Foster, R.C., et al. Pediatrics 73(6), 879 (1984).
- Jones, J.E., Causey, C.P., Knuckley, B., et al. Curr. Opin. Drug Discov. Devel. 12(5), 616-627 (2009).
- Schellekens, G.A., de Jong, B.A., van den Hoogen, F.H., et al. J. Clin. Invest. 101(1), 274-281 (1998).
- Conrad, K., Roggenbuck, D., Reinhold, D., et al. Autoimmun. Rev. 9(6), 431-435 (2010).
- Schellekens, G.A., Visser, H., deJong, B.A., et al. Arthritis Rheum. 43(1), 155-163 (2000).
- Wiik, A.S., van Venrooij, W.J., and Pruijn, G.J.M. Autoimmun. Rev. 10(2), 90-93 (2010).
- Duskin, A. and Eisenberg, R.A. Immunol. Rev. 233(1), 112-125 (2010).
- Wang, Y., Wysocka, J., Sayegh, J., et al. Science 306(5694), 278-283 (2004).
- Cuthbert, G.L., Daujat, S., Snowden, A.W., et al. Cell 118(5), 545-553 (2004).
- Lee, Y.-H., Coonrod, S.A., Kraus, W.L., et al. Proc. Natl. Acad. Sci. USA 102(10), 3611-3616 (2005).
- Denis, H., Deplus, R., Putmans, P., et al. Mol. Cell Biol. 29(18), 4982-4993 (2009).
- Métivier, R., Penot, G., Hübner, M.R., et al. Cell 115(6), 751-763 (2003).
- Li, P., Yao, H., Li, M., et al. Mol. Cell Biol. 28(15), 4745-4758 (2008).
- Li, P., Wang, D., Yao, H., et al. Oncogene 29(21), 3153-3162 (2010).
- Chang, X., Han, J., Pang.L, et al. [In Press] BMC Cancer (2009).
- Slack, J.L., Jones, L.E., Jr., Bhatia, M.M., et al. Biochemistry 50(19), 3997-4010 (2011).
- Brinkmann, V., Reichard, U., Goosmann, C., et al. Science 303(5663), 1532-1535 (2004).
- Wang, Y., Li, M., Stadler, S., et al. J. Cell Biol. 184(2), 205-213 (2009).
- Li, P., Li, M., Lindberg, M.R., et al. J. Exp. Med. 207, 1853-1862 (2010).